 The background of major quality traits in soybean
The background of major quality traits in soybean
Abstract
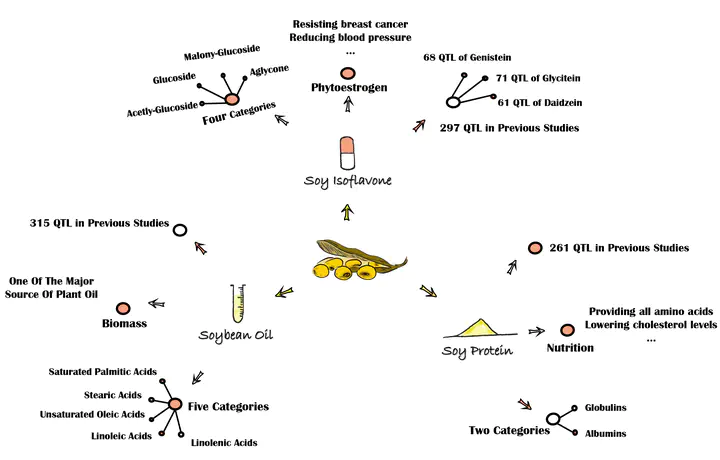
Isoflavone, protein, and oil are the most important quality traits in soybean. Since these phenotypes are typically quantitative traits, quantitative trait locus (QTL) mapping has been an efficient way to clarify their complex and unclear genetic background. However, the low-density genetic map and the absence of QTL integration limited the accurate and efficient QTL mapping in previous researches. This paper adopted a recombinant inbred lines (RIL) population derived from ‘Zhongdou27’and ‘Hefeng25’ and a high-density linkage map based on whole-genome resequencing to map novel QTL and used meta-analysis methods to integrate the stable and consentaneous QTL. The candidate genes were obtained from gene functional annotation and expression analysis based on the public database. A total of 41 QTL with a high logarithm of odd (LOD) scores were identified through composite interval mapping (CIM), including 38 novel QTL and 2 Stable QTL. A total of 660 candidate genes were predicted according to the results of the gene annotation and public transcriptome data. A total of 212 meta-QTL containing 122 stable and consentaneous QTL were mapped based on 1,034 QTL collected from previous studies. For the first time, 70 meta-QTL associated with isoflavones were mapped in this study. Meanwhile, 69 and 73 meta-QTL, respectively, related to oil and protein were obtained as well. The results promote the understanding of the biosynthesis and regulation of isoflavones, protein, and oil at molecular levels, and facilitate the construction of molecular modular for great quality traits in soybean.